Rationale – Background and training in bioinformatics tools is required for the research community at UCSF to remain at the forefront of biomedical research and successfully compete for funding. Bioinformatics tools help translate our collective molecular understanding of disease into actionable insights and life-improving patient care innovations. Application of bioinformatics knowledge can minimize persistent barriers to progress in the translational research workflow by providing a bridge between the domain expertise of the experimentalist and the world of computationally driven, information-based research methods (see Appendix Fig 1).
Currently at UCSF, training on bioinformatics tools is decentralized. Services, databases, and courses are hosted by Core Labs, the UCSF Library, and other departments. Centers of data generation are not always integrated with the tools needed to gain insight from the data or place data in a disease-context. Navigating lists of bioinformatics tools can be a daunting task for researchers with no background in bioinformatics (see Appendix for current lists).
This arrangement carries numerous risks:
- Insufficient information about the best tools and platforms to address a particular research question.
- Inability to accurately gauge the true investment needed in both time and money to gain full value from an experiment.
- Poor return on investment from expensive experiments, and missed opportunities due to lack of awareness about tools available to translate raw data into findings and testable hypotheses.
In keeping with the UCSF Library’s commitment to support the continuing educational needs of information resource users, and extend services to a wider audience of researchers, we propose a pilot project to evaluate web-based methods for bioinformatics training. The UCSF Library will provide access to a centralized, web-based resource for training on well-adopted, authoritative bioinformatics resources by hosting the OpenHelix collection of bioinformatics tutorials on its website. This new service would cross-reference with related services (Core Labs, Cores Search, MyCORES etc.) to provide centralized and coordinated access to bioinformatics tools training at UCSF.
We are aware that there are a number of different groups at UCSF with expertise and interest in bioinformatics training and our aim with this proposal is to work with these groups to identify needs and develop solutions for translational medicine researchers.
Successful implementation of this new bioinformatics training service can:
- Provide researchers with the skills needed to locate, learn about, and apply the information housed in bioinformatics databases.
- Enable them to identify the right tool for their particular research question.
- Improve collaborations between experimentalists and bioinformatics experts at UCSF.
- Reduce time between data generation and insight by making bioinformatics training and tools available to those who generate data at the time it is generated.
- Reduce the current burden on Core Labs that may not have time for basic bioinformatics training requests from researchers.
- Establish an engaged user base through which the UCSF Library and Research Resource Program can continually evaluate services, identify unmet needs for training and resources, and design new services.
Plan
- Collaborate with the UCSF Institute for Computational Health Sciences, leveraging their expertise in bioinformatics to identify researchers' training needs and develop solutions to meet those needs.
- Form a Bioinformatics Tools User Group (user base for this pilot)
- Survey user group to identify bioinformatics tools and training needs
- Evaluate OpenHelix as an inexpensive, out of the box solution for bioinformatics training
- Make Go/No Go decision on value of OpenHelix
- Design and implement library-based home for resources such as OpenHelix: Bioinformatics @UCSF Library
- Collaborate with CTSI to create Marketing and Outreach plan for successful adoption of new service
- Ongoing evaluation of web-based bioinformatics training service and development of a business model for continued support of the service.
Criteria and Metrics for Success:
- Pilot user base includes bioinformatics subject matter experts as well as end users.
- Good response rates on User-Needs and OpenHelix evaluation surveys
- Steady increase in hits to library-based bioinformatics website 6 months after launch
- Steady increase in utilization of OpenHelix tutorials 6 months after launch
- Good response rate on User Satisfaction survey 1 year after launch of new service. Metrics identifying most/least utilized bioinformatics tools.
- Increase in User Satisfaction rating 2 years after launch of new service
Collaborators:
Julie Auger, Executive Director of UCSF Research Resource Program (RRP)
Budget includes funding for:
- 2-year license to OpenHelix
- Programming resources to integrate with existing web infrastructure (UCSF Library, Core Labs and Cores Search)
- Minimal in-kind CTSI support for survey development, marketing and outreach plan
- External trainers for on-site training on most highly valued bioinformatics tools.
APPENDIX
1. Hyperlinks to current lists of bioinformatics tools
http://gettinggeneticsdone.blogspot.com/2011/06/resources-for-pathway-analysis.html
http://grouthbio.com/Genome_Software_Service.php
http://www.oxfordjournals.org/nar/database/a/
http://www.hsls.pitt.edu/obrc/
2. Examples of Medical Libraries that host or are evaluating OpenHelix as part of their library-based bioinformatics services
- Mt. Sinai School of Medicine Levy Library –successful implementation with 5-year renewal and fully booked on-site training classes
- Becker Medical Library – Wash U
- University of Illinois
- Emory – Woodruff Health Sciences
- University of Pittsburg
- Weill Cornell Medical Library
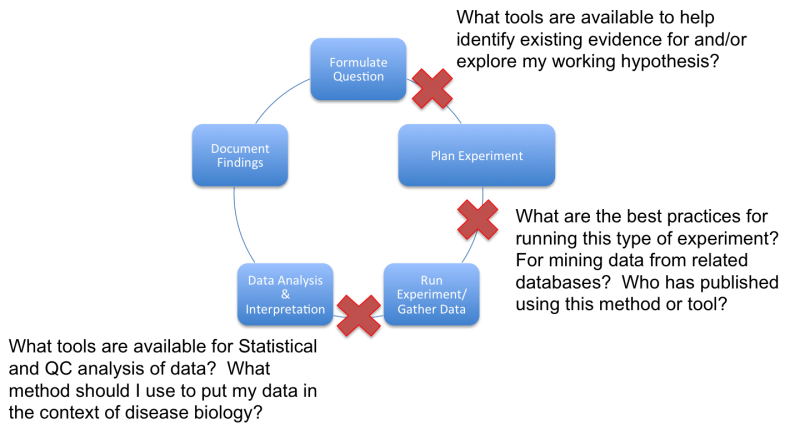
3. Figure 1. Awareness and training in bioinformatics tools can eliminate persistent roadblocks in the translational research workflow.
Commenting is closed.

Comments
This is great idea! UCSF and
This is great idea! UCSF and Gladstone are top notch research institutions, yet in an era when Big Data permeates the research culture, we lack a cohesive plan for building stronger bioinformatics infrastructure and training opportunities. There is considerable bioinformatics and computational biology talent on campus and this proposal could help bring together that talent to develop and implement such a plan. Count me in!
As executive director of the
As executive director of the National Resource for Network Biology (NRNB) and member of UCSF Mission Bay campus, I strongly support this proposal. The current state of training for bioinformatics tools and methods is disorganized and would greatly benefit from the proven leadership of UCSF Library's CKM. We have a growing collection of tutorial materials for network biology tools, such as Cytoscape and WikiPathways, as well as local expertise and presenters, which we would contribute to a centralized training program. Let us know *when* this worthy endeavour is funded and launched!
Thanks Alisha and Alex for
Thanks Alisha and Alex for your comments. We see the OpenHelix resource as a great way to jump start this effort and get a critical mass of tutorials avilable to the UCSF research community quickly, and in a pretty cost-effective way. The Library-based home for this training resource could then be optimized with links to advanced tutorials from in-house experts, announcements about upcoming training & seminars, etc, which would complement the basic tutorials OpenHelix has for resources such at Cytoscape and GenMAPP.
This is an excellent idea,
This is an excellent idea, but for CTSI funding, we have to make sure that the the program covers the CTSI National Consortium Education and Career Development Key Function Committee required competencies for bioinformatics. In addition, this effort should be coordinated with the CTSI Biomedical Informatics Program training activities.
Thanks for your comment
Thanks for your comment Deborah.
Our interest in Open Helix as a possible source for training material comes from the desire to quickly address an immediate need that probably falls under the general Biomedical Informatics core competency theme, that is - to help researchers understand the role of bioinformatics in study design and analysis of genomic, proteomic, and metabolic data, and also get hands on experience with the most commonly used computational tools, bioinformatics databases, and data repositories to help them formulate research questions. Designing these tutorials from scratch would be outside the scope of our existing resources.
Yes, we definitely would want to coordinate this with CTSI Biomedical Informatics Program training activities. A library-based portal for bioinformatics training would be a great place to disseminate any educational materials (recorded webinars, tutorials, hands on exercises) created by the CTSI Biomedical Informatics Program, and to promote bioinformatics training events and courses that occur throughout UCSF.
This resource could also be a way to increase awareness of and provide training on bioinformatics tools created through CTSA funding such as openSESAME, as well as tools (and experts) needed for biostatistical analysis of data such as SAS and SPSS - software that is available through the Research Software Licensing arm of the library.
Thanks for your interest in the proposal. I'll also contact you via e-mail for advice on resources for getting up to speed on CTSI Key Function Committees.
Overall, I think this is a
Overall, I think this is a great initiative, but it needs to be coupled with hands-on courses offered either through the library or facilitated by the library. One of the major challenges we have in offering training on Cytoscape, Chimera, or other bioinformatics tools is management of the logistics. In the past, we have worked with the library to offer a summer series on Chimera (see http://www.cgl.ucsf.edu/Outreach/Workshops/ at the bottom of the page) that was well attended (most sessions were completely subscribed). I think OpenHelix might be a way to gain interest, but I think that to serve the deep needs of the UCSF community it needs to be coupled with hands-on courses.
Scooter Morris
Executive Director, NIH Resource for Biocomputing, Visualization, and Informatics
Director, Sequence Analysis and Consulting Service
Hi Scooter. Thanks for
Hi Scooter. Thanks for taking the time to read through the proposal and for your comment. I completely agree. Hands-on courses and online training are very complementary in nature and this program could facilitate both. The self-serve training model provided by online tutorials and exercises can fill an on-demand need for education and training, at the time that researchers need it. But the level of training researchers require in order to understand how to apply these tools and databases to their specific research question requires hands-on training and examples from their peers and experts in the field.
We have some really good momentum going right now with the Pathway Analysis workshops offered through the UCSF Library (oversubscribed at the moment), but the focus of those workshops has been on a single tool for analysis of 'omics data (network analysis, functional analysis, pathway enrichment). Now would be a great time to build on that momentum and line up some additional training/courses on tools and databases that are already in demand here at UCSF. I think that is an effort we would like to work with you and other groups on (e.g. the Gladstone Bioinformatics group) regardless of whether this proposal is funded.
Adding to the bioinformatics training toolkit with a library of on-demand/self-serve tutorials can fill in the gaps when researchers have that immediate need. And any materials (recorded webinars, slides, etc.) from hands-on courses from UCSF experts would find a home there as well.
Thanks again for your suggestion. I'll contact you off-line about courses and training you would recommend in the short term. The library is very well suited to facilitate those courses.