1. The UCSF Health Problem
Prostate cancer is the most commonly diagnosed malignancy among men in the United States. Accurate diagnosis, staging, and monitoring are critical for effective treatment planning and improving patient outcomes. Imaging modalities, such as Prostate-Specific Membrane Antigen (PSMA) Positron Emission Tomography (PET), CT, and MRI, provide valuable insights but often lack the integration necessary to fully capture the complexity of prostate cancer progression. Current challenges include:
- Fragmented Data Interpretation: Clinicians must manually synthesize information from disparate imaging modalities (PSMA PET, CT, MRI) and clinical records, which can be time-consuming and prone to variability.
- Variability in Imaging Assessments: Interpretations of imaging studies can vary among radiologists, leading to inconsistencies in diagnosis and treatment planning.
- Diagnostic Workflow Inefficiency: Radiologists face growing workloads with increasingly complex multimodal cases. Generating comprehensive, standardized reports is labor-intensive and varies by provider.
AI methods have exceptional potential to better leverage the full spectrum of available data, including imaging, and clinical measures, to improve performance. The intended end-users of this project are medical oncologists, radiation oncologists, radiologists, and urologists, who currently lack integrated tools to synthesize these multimodal data.
2. How might AI help?
A multimodal foundation model leveraging PSMA PET/CT/MRI datasets and textual clinical data from UCSF prostate cancer patients offers a transformative solution. PSMA PET has recently emerged as an extremely powerful tool for more accurately identifying prostate cancer cells, particularly in the metastatic setting. It is combined with CT and/or MRI to provide anatomical reference, allowing for identification of localization patterns (e.g. metastases to the bones vs liver vs lymph nodes) as well as removing false positives. MRI has the additional benefit of potentially incorporating additional contrasts depicting perfusion and cell density that also can reveal tumor characteristics. Complementing this, textual clinical data encompasses a wealth of information regarding patient history, symptoms, PSA levels, biopsy results (including Gleason scores), treatment regimens, and follow-up outcomes. By integrating imaging and clinical data, a multimodal vision foundation model can potentially uncover intricate relationships and patterns that remain hidden when each data type is analyzed separately. Furthermore, a significant advantage of multimodal foundation models lies in their ability to learn from large-scale unlabeled or naturally paired data, which is particularly beneficial given the challenges and costs of annotating large, multimodal datasets.
Phase 1 will focus on developing a pre-trained multimodal foundation model using a substantial corpus of PSMA PET/CT/MRI images and corresponding textual clinical data from prostate cancer patients at UCSF. We have a PSMA PET database of over 2000 studies at UCSF which will be used to identify the datasets. This will be done with self-supervised learning. Phase 2 will involve fine-tuning the pre-trained foundation model for two downstream high-impact applications that directly assist clinical workflow:
- Lesion Detection & Segmentation: Automate identification and outlining of cancerous lesions on scans, to assist in consistent tumor detection.
- AI-Generated Radiology Reports: Automatically produce a draft radiology report from the multimodal data, reducing the reporting workload on radiologists.
By project end, we will deliver (1) a pre-trained multimodal foundation model, (2) a fine-tuned lesion segmentation AI tool, and (3) a fine-tuned report generation AI tool validated on UCSF data.
3. How would an end-user find and use it?
When deployed, clinicians will access the AI tool through familiar systems (the APeX EHR and PACS imaging viewer). For example, when a prostate cancer patient’s PSMA PET/CT or MRI is opened, an AI Decision Support panel will display the model’s outputs. These include visual lesion overlays on the scans (highlighting detected tumors or metastases) and an AI-generated draft report summarizing key findings (e.g. lesion locations and characteristics). The radiologist can review the highlighted lesions, adjust or accept the draft text, and be notified of any critical findings flagged by the AI. Explanations (e.g. confidence levels or reference images for each prediction) will accompany the results to help the clinician trust and understand the recommendations. This AI support is embedded seamlessly into the existing workflow via a tab in APeX/PACS, so end-users can incorporate the tool without needing to switch to a separate application.
4. Embed a picture of what the AI tool might look like.
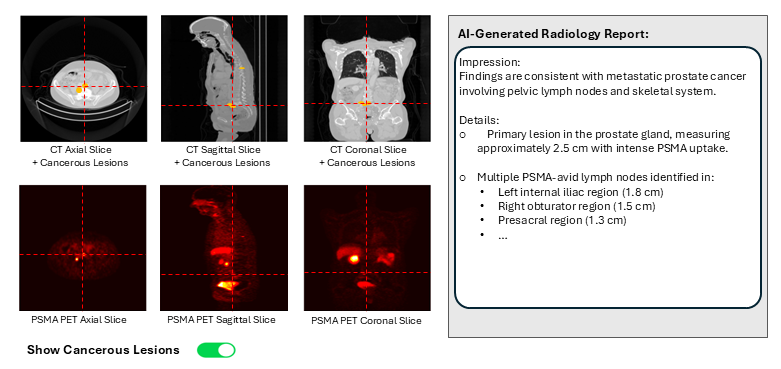
Figure: AI Decision Support interface for prostate cancer care, showing multimodal imaging (CT and PSMA PET shown) with cancerous lesion detection and classification overlays, and an auto-generated report summarizing the findings is displayed.
5. What are the risks of AI errors?
Risks include false positives (benign findings flagged as cancer), false negatives (missed cancer lesions), hallucinations (incorrect information in the report), algorithmic bias (disparities in care), and overreliance on AI (overlooking clinical judgment). We will measure error rates by comparing AI predictions to ground truth data (histopathology, outcomes) and through clinician feedback. Mitigation strategies include using high-quality training data, rigorous validation, uncertainty estimation, transparent presentation of limitations, human oversight and clinician override options. Continuous monitoring and retraining will be essential.
6. How will we measure success?
a. Using Existing APeX Data:
- Diagnostic Accuracy: Comparison of AI-assisted diagnoses with traditional methods using metrics such as sensitivity, specificity, and area under the curve (AUC).
- Workflow Efficiency: Time taken for report generation and decision-making processes before and after AI implementation.
- Number of unique clinicians using the tool and frequency of use.
b. Additional Measurements:
- Surveys and feedback from clinicians on usability and clinical utility.
- Prospective evaluation of AI accuracy in downstream tasks (e.g., checking segmentation accuracy and report correctness in a pilot setting).
Continued support from UCSF Health leadership will require demonstrating significant clinician adoption, positive feedback, improvements in intermediate outcomes or workflow efficiencies, and evidence of the AI model's accuracy.
7. Describe your qualifications and commitment:
I am Mansour Abtahi, a Specialist at the University of California San Francisco, where I am currently developing AI/ML models on prostate cancer using clinical and imaging data (PET/CT/MRI). I bring extensive expertise in developing innovative AI/ML models within healthcare, as evidenced by my publications on analyses of ophthalmology imaging data in top-tier journals. My skills encompass a wide range of deep learning architectures, including CNNs, Transformers, ViTs, and Large Multimodal Models, along with proficiency in computer vision techniques for medical image analysis, particularly in 3D imaging (MRI, CT, PET) relevant to this project. I am well-positioned to co-lead with esteemed UCSF faculty such as Dr. Thomas Hope and Dr. Peder Larson. Dr. Hope, Vice Chair of Clinical Operations in Radiology, has led the translation of PSMA PET imaging into the clinic, creating a paradigm shift in prostate cancer assessment, and the use of theranostic agents for precision treatment of prostate cancer. Dr. Larson, Director of the Advanced Imaging Technology Research Group, is a biomedical engineer who specializes in imaging physics and multimodality imaging, with projects improving prostate cancer imaging with PET/MRI and hyperpolarized metabolic MRI. Collaborating with them aligns perfectly with the goals of this project. I am fully committed to dedicating up to 10% of my time as a co-lead, participating in progress sessions, and collaborating closely with UCSF’s Health AI and AER teams. This effort will drive the successful integration of AI models into the APeX EHR system, with the goal of improving prostate cancer patient outcomes at UCSF Health.
Comments
Sounds like a good project.
Sounds like a good project. Do you have gold standard annotated images to use for training the model to detect and segment lesions? Is 2000 images enough to build a strong foundation model?
Thank you. Yes, we have over
Thank you. Yes, we have over 200 annotated images for lesion detection and segmentation.
While more data is always helpful, based on other successful studies, the 2,000 cases we have should be sufficient to train a strong foundation model, especially using self-supervised learning.
I’ve added two relevant
I’ve added two relevant studies involving PET/CT and textual datasets for reference.
Yujin et al., Developing a PET/CT Foundation Model for Cross-Modal Anatomical and Functional Imaging, arXiv:2503.02824 (2025).
Wang et al., An Innovative and Efficient Diagnostic Prediction Flow for Head and Neck Cancer: A Deep Learning Approach for Multi-Modal Survival Analysis Prediction Based on Text and Multi-Center PET/CT Images. Diagnostics 14, no. 4 (2024): 448.